Introduction
Multiple myeloma (MM) is a cancer of plasma cells characterized by increased survival and proliferation of terminally differentiated plasma cells in the bone marrow.1 Clinically, MM diagnosis is prompted by the detection of monoclonal intact immunoglobulin (M spike), also known as free light chains (not in association with heavy chains), in the serum and urine of patients presenting with 1 or more of hypercalcemia, renal failure, anemia, and bone disease (the CRAB criteria). Major progress in our understanding of MM biology over the past 4 decades has led to significant improvements in how we treat MM, reflected by a 3- to 4-fold increase in patient median survival. Although MM is now better controlled over longer periods for many patients, it remains incurable and resistance to novel agents represents a major clinical problem. This review will focus on the molecular mechanisms underlying protein handling in MM and on bench-to-bedside translation of therapies targeting protein synthesis, folding, and degradation in MM. Rational combination of these agents holds promise to help overcome proteasome inhibitor (PI) resistance in MM, with the goal of achieving prolonged remission, if not cure, for most patients with multiple myeloma.
Scientific Rationale for Targeting Protein-Handling Pathways in Multiple Myeloma
The process of protein synthesis and folding is intrinsically prone to errors, and eukaryotic cells are equipped with quality control mechanisms to ensure that native proteins adopt proper tertiary and quaternary conformations. The cytotoxic accumulation of misfolded proteins causes endoplasmic reticulum (ER) stress and activates the unfolded protein response (UPR), which, together with autophagy, aggresome, and the ubiquitin-proteasome system (UPS), has the goal of maintaining protein homeostasis.2,3 It is estimated that one-third of newly synthesized proteins are de- graded via the proteasome within minutes of their synthesis dueto an intrinsic inability to achieve stably folded conformations.4 These rapidly degraded proteins are termed “defective ribosomal products” (DRiPs). Due to high protein turnover, cancer cells typically produce an even higher percentage of DRiPs, making them reliant on an intact UPS for survival.5-8 This is especially true for MM, a cancer characterized by a high synthesis rate of immunoglobulins. In fact, MM cells exhibit stigmata of ongoing proteotoxic stress with baseline induction of UPR and accumulation of polyubiquitinated proteins, providing a substrate for proteasome-mediated degradation.9-11
Studies have shown that an imbalance between the cargo for proteasomal degradation (polyubiquitnated proteins) and the activity of the proteasome is a key determinant of PI sensitivity in MM.12 Drugs that increase proteasome workload (eg, heat shock protein [HSP] inhibitors, ER stressors) synergize with drugs that decrease proteasome activity (eg, PIs) in MM. The results of in vitro studies have shown that proteasome inhibition, perhaps even UPR induction, results in the compensatory activation of aggresome, autophagy, and heat shock response pathways in an effort to protect MM cells from proteotoxicity (Figure 1).2,13-15 Further work assessing the combinatorial effects of blocking 2 or more of these pathways in MM are currently ongoing, some of which are highlighted below.
Ubiquitin-Proteasome System
At the core of protein homeostasis in eukaryotes is the UPS (Figure 1A). Proteins targeted for proteasome degradation are polyubiquitinated via a 3-enzyme cascade involving E1 (activating), E2 (conjugating), and E3 (ligase) enzymes, while deubiquitinating enzymes (DUBs) act in opposition to E3 ligases to remove ubiquitin.16-19 The 26S proteasome is an ATP-dependent, multicatalytic complex comprising a 20S catalytic core flanked on either side by 19S regulatory caps.20 Polyubiquitinated substrates are recognized by the 19S regulatory subunit that, in concert with DUBs, remove ubiquitin and facilitate engagement with the 20S core that contains the catalytically active β1 (caspase-like activity), β2 (trypsin-like activity), and β5 (chymotrypsin-like activity) subunits.19,21,22 The PIs bortezomib, carfilzomib, ixazomib, and oprozomib primarily target the β5 subunit, while marizomib appears to have activity against all 3 β-subunits.23-26
Prior to degradation, proteasome-associated DUBs (eg, RPH11, UCH37, and USP14) remove ubiquitin chains, which would otherwise sterically hinder the translocation of target proteins to the 20S core.27 Similar to PIs, DUB inhibitors trigger poly- ubiquitinated protein accumulation and apoptosis in MM, but without inhibiting the catalytic subunits of the proteasome.28-30 Thus, DUB inhibitors could theoretically overcome resistance to proteasome inhibition when this is mediated by mutations in the catalytic subunits of the proteasome. Furthermore, DUB inhibition offers the opportunity to promote the degradation of proteins that are preferential clients of specific DUBs.
Autophagy
Autophagy, a conserved process of autoproteolysis that plays a key role in maintaining protein homeostasis (Figure 1B), participates in the quality control of protein synthesis/degradation by sequestering misfolded/aggregated proteins in autophagosomal vesicles for subsequent lysosome degradation.31 Studies have shown that crosstalk exists between UPS, ER stress, and autophagy.32-34 Although elevated basal autophagic activity in primary MM cells is associated with shorter overall survival (OS) and progression-free survival (PFS), autophagy’s role in MM is controversial given that it can be pro-survival and pro-apoptotic depending on factors we have yet to fully understand.32 The current consensus is that a basal level of autophagy is essential for MM survival as an alternative proteolytic pathway in the face of decreased proteasome activity/increased proteotoxic stress, thus providing a rationale for the combination of autophagy inhibitors with PI in MM. However, persistent, sustained, and uncontrolled autophagy is likely to result in cell death, outlining the difficulties in therapeutically targeting autophagy.35
Aggresome Pathway
In vitro, the aggresome pathway is activated when proteasomes are blocked. Polyubiquitinated protein aggregates are transported along the microtubule to the microtubule-organizing center in a histone deacetylase 6 (HDAC6)-dependent manner to form aggresomes that target proteins for refolding or degradation by autophagy (Figure 1C).36-39 The results of in vitro studies show that combined inhibition of the proteasome and aggresome leads to synergistic cell death in MM, providing strong rationale for combining PI with HDAC6 selective inhibitors.40
Heat Shock Chaperone Proteins
Heat shock chaperone proteins (HSPs) are a class of enzymes that chaperone the proper folding and function of proteins, and that direct misfolded proteins to degradation; therefore, they participate in protein quality control (Figure 1D).41-44 In MM, HSPs support proliferation and survival by 1) facilitating proper folding of newly synthesized proteins to prevent proteotoxic stress and 2) preferentially supporting the folding and expression of several oncogenes.45 Two main families of HSPs are being targeted therapeutically in MM: HSP90 and HSP70. Interestingly, HSP70 overexpression in neurons results in inhibition of caspase-dependent and -independent apoptosis, suggesting a third pro-survival function.46,47 Inhibition of HSP90 or the proteasome results in compensatory upregulation of HSP70, thereby making the latter an attractive target in combinatory anti-MM therapy. Recently, there has been growing interest in developing inhibitors against heat shock factor 1 (HSF1), the “master regulator” of heat shock response, in an attempt to avoid compensatory upregulation of individual chaperones.45
Endoplasmic Reticulum Stress and Unfolded Protein Response
The UPR is a tripartite response triggered by the accumulation of unfolded/misfolded proteins in the ER (Figure 1E).48,49 The UPR functions to restore equilibrium in the ER; however, prolonged/ persistent activation of UPR results in apoptosis.50 The 3 distinct UPR branches are regulated by 3 kinases: IRE1, PERK, and ATF6. Activation of IRE1 results in the splicing of XBP1 mRNA which, together with activated ATF6, regulates ER expansion, increases expression of chaperone proteins, and initiates ER-associated de-gradation to reduce ER stress.51 Depending on the magnitude and duration of stress, IRE1 can either activate antiapoptotic signaling through protein kinase B or trigger apoptosis through c-Jun N-terminal kinases (JNK) activation.52-54 Further- more, JNK activation can initiate autophagy, thereby serving as a link between ER stress and autophagy.54 Finally, protein kinase R-like ER kinase (PERK) activation inhibits eIF2α, leading to a repression of global protein synthesis while selectively inducing the translation of ATF4.55 ATF4 activates cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP), and together they upregulate protective autophagy in the face of transient proteotoxic stress.56 However, if the stress is prolonged, CHOP can trigger apoptosis, outlining the double-edged nature of this stress response pathway.57
Forced expression of spliced X-box binding protein 1 (XBP1) in B cells induces an MM-like phenotype in mice, and high XBP1 expression in primary MM cells correlates with poor OS, suggesting that chronic IRE1-XBP1 activation may be important for MM survival.58 However, it was recently demonstrated that decreased XBP1 splicing confers bortezomib resistance in MM.59 By suppressing XBP1s, MM cells decommit to plasma cell maturation and decrease immunoglobulin production, proteasome load, and ER stress, resulting in acquired resistance to PI.12,59
Clinical Translation of Therapies Targeting Protein-Handling Pathways in Multiple Myeloma
Drugs Targeting the Ubiquitin-Proteasome Pathway
Apart from FDA-approved bortezomib, carfilzomib, and ixazomib, there are 2 novel PIs in advanced clinical development, oprozomib and marizomib. Oprozomib (ONX 0912), an oral analogue of carfilzomib, is an irreversible epoxyketone PI. In preclinical studies, oprozomib demonstrated cytotoxicity in MM in combination with lenalidomide and/or HDAC inhibitor molecules, as well as bone anabolic effects.60,61 A phase Ib/II trial of single-agent oprozomib showed an overall response rate (ORR) of 22% to 34% in relapsed/refractory MM (R/R MM), including bortezomib- and carfilzomib-refractory MM (NCT01416428).62 In the R/R MM setting, the combination of oprozomib with dexamethasone (NCT01832727) or with pomalidomide and dexamethasone (NCT01999335) has resulted in ORRs of 42% and 50% to 59%, respectively.63,64
Attempts at overcoming PI resistance by blocking all proteasome catalytic subunits prompted the development of marizomib, a pan-proteasome inhibitor.65,66 A phase I study of marizomib (NCT00461045) reported an ORR of 7.4% in bortezomib-, lenalidomide-, and/or thalidomide-refractory patients.67 Based on encouraging preclinical data supporting the combination of marizomib and pomalidomide/dexamethasone, clinical trials evaluating this combination are now underway (NCT02103335).68
Deubiquitinating Enzyme InhibitorsThe small-molecule DUB inhibitors RA190, P5091, and B-AP15 target RPN13, USP7, and USP14/UCHL5, respectively. In vitro, they induce proteotoxicity and apoptosis in MM without direct inhibition of catalytic subunits of the proteasome.69-71 The USP14 inhibitor VLX1570 also demonstrated promising preclinical activity and is currently being evaluated in an early-phase clinical trial in MM.28
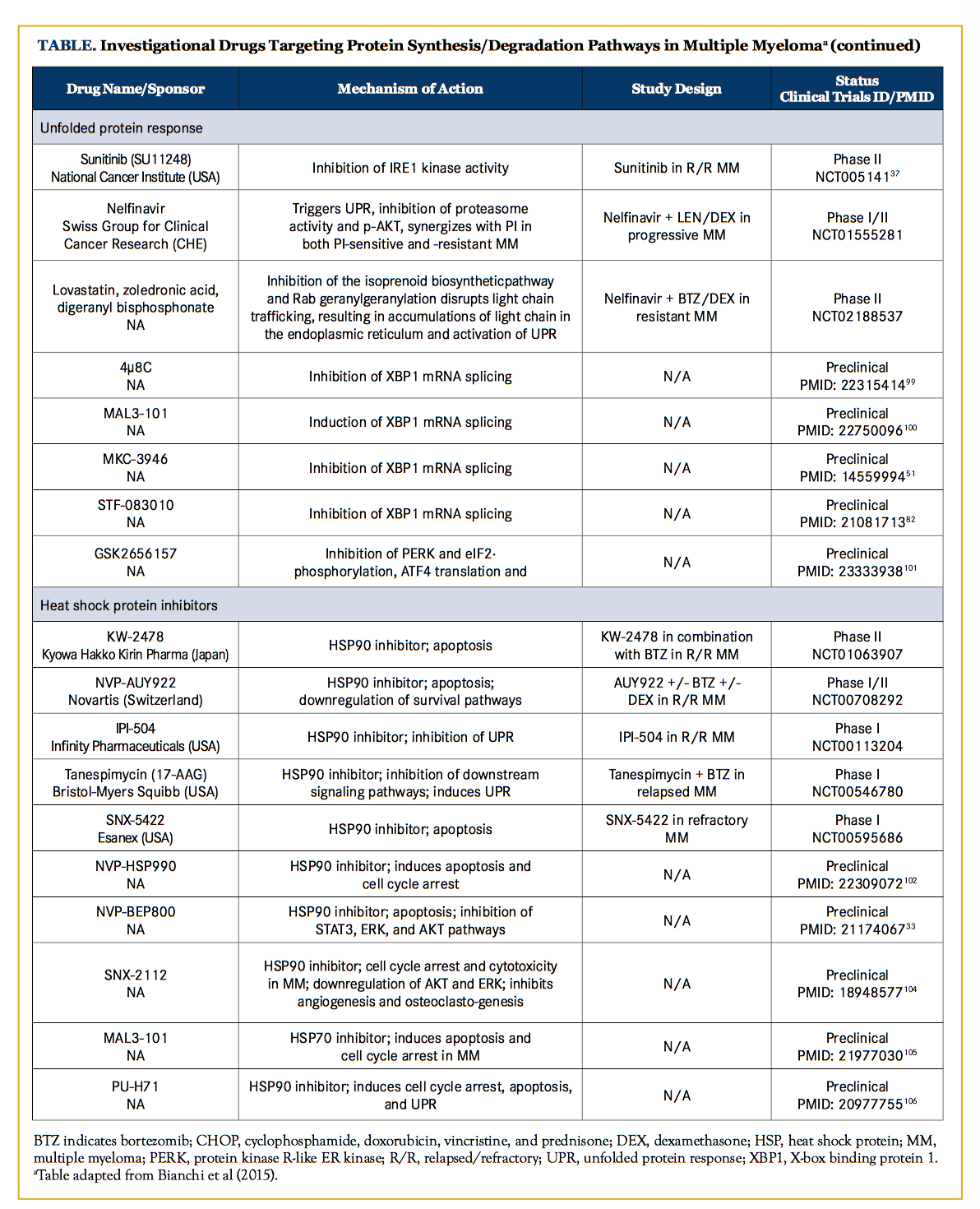
Heat Shock Chaperone Protein Inhibitors
HSP90 is the most well-studied chaperone protein in MM. Several HSP90 inhibitors have completed phase I clinical studies (Table). Notably, among patients who were evaluable (n = 67), the combination of tanespimycin with bortezomib in R/R MM was associated with an ORR of 15%.72 However, modest activity and/ or significant toxicity hampered clinical development of next-generation HSP90 inhibitors.73
Histone Deacetylase Inhibitors
Panobinostat, in combination with bortezomib and dexamethasone, was recently approved as a third-line therapy in patients with MM with prior bortezomib and immunomodulatory drug (IMiD) exposure. Vorinostat is a class I and II HDAC inhibitor currently undergoing clinical trials. The phase III Vantage 008 trial reported improvements in ORR (54% vs 41%; P <.0001) and PFS (7.6 vs 6.8 months; P = .10) when vorinostat was added to bortezomib.74 Although nonselective HDAC inhibitors show promising clinical activity, they are associated with significant adverse effects (particularly of a gastrointestinal and hematologic nature) due to the indiscriminate targeting of multiple HDACs, leading to extensive modulation of downstream histone and nonhistone protein functions.75 Isoform-specific HDAC inhibitors, focusing on inhibition of HDAC6 and the aggresome pathway, have been developed in an effort to maintain efficacy and limit toxicities. Two promising HDAC6-selective inhibitors (ACY-241 and ACY-1215) are currently undergoing clinical trials. A phase Ib study of ricolinostat (ACY- 1215) in combination with bortezomib/dexamethasone reported 45% and 25% ORRs in R/R MM and bortezomib-refractory MM, respectively, while ricolinostat in combination with lenalidomide/ dexamethasone had an ORR of 55% in R/R MM.76,77 BG45, an HDAC3-selective inhibitor, has also shown promising preclinical results, demonstrating anti-myeloma activity alone and in combination with bortezomib; translation to an early phase clinical trial is anticipated soon.78
Unfolded Protein Response Modulators
Pharmacological induction of UPR via ER stressors, such as tunicamycin, thapsigargin, and brefeldin A, has proven to potently synergize with a PI in vitro; however, clinical translation is limited by anticipated toxicities based on animal models.11,79-81 A more elegant way to exploit the UPR against MM hinges on identifying which of the tripartite signaling pathways and their downstream effectors are necessary for MM survival and proliferation. Such identification efforts were recently undertaken, revealing that single knockdown of each branch of the UPR has modest effects on MM viability at baseline; however, an intact IRE1-XBP1 pathway is required for bortezomib-mediated cytotoxicity.59 Inhibition of either the endoribonuclease or kinase domains of IRE1 was found to have an anti-MM effect, especially combined with PI.82,83
PERK inhibition has also recently emerged as a potential therapeutic option in MM. In preclinical studies, GSK2606414, a selective PERK inhibitor, was shown to synergistically enhance the apoptotic effect of bortezomib in MM.84 Nelfinavir is an HIV-protease inhibitor that has demonstrated anti-MM activity through UPR induction, CHOP upregulation, poly (ADP-ribose) polymerase cleavage, and proteasome inhibition in preclinical studies.85 Nelfinavir could also re-sensitize bortezomib-refractory primary MM cells towards bortezomib treatment.85 A phase II trial of nelfinavir and bortezomib reported an ORR of 30% in the dose escalation cohort and an ORR of 50% in an exploratory extension cohort comprising patients with both bortezomib-refractory and lenalidomide-resistant MM.86 No clinical-grade inhibitors of ATF6 have been reported at the time of writing.
Immunomodulatory Drugs
The clinical synergism between proteasome inhibitors and the IMiDs thalidomide, lenalidomide, and pomalidomide is well established.87,88 The molecular mechanisms of activity of these compounds have long been obscure, and the anti-angiogenic effect was initially thought to be primarily responsible for their anti-MM activity.89 However, the degradation of the transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) was recently shown to be the base of the anti-MM effect of lenalidomide.90,91 In an unexpected twist, lenalidomide was shown to bind to the E3 ubiquitin-ligase complex made up of the damage-specific DNA-binding protein 1 (DDB1) and cereblon, enhancing its activity and facilitating ubiquitination and proteasome-mediated degradation of IKZF1 and IKZF3. IMiD-mediated stimulation of thymus and natural killer (NK) immunity similarly depends on the degradation of IKZF1 and IKZF3, resulting in IL-2 production in T lymphocytes. Based on these findings, the clinical synergism between bortezomib, a PI, and lenalidomide, a facilitator of proteasome-mediated IKZF1 and IKZF3 degradation, appears paradoxical and remains to be clarified at the cellular and molecular levels.
Conclusions and Future Directions
Although disrupting proteostasis via a PI has been successful in MM, innate or acquired resistance remains a major clinical challenge. Combination treatments have only partially overcome these issues, and progressive acquisition of resistance to multiple agents with each disease relapse is a well-known phenomenon in MM. Recent research efforts have focused on modulating other facets of protein homeostasis pathways (ie, aggresome, autophagy, UPR, DUB, HSP), with the goal of exacerbating proteotoxcity and overcoming MM drug resistance (Figure 1).
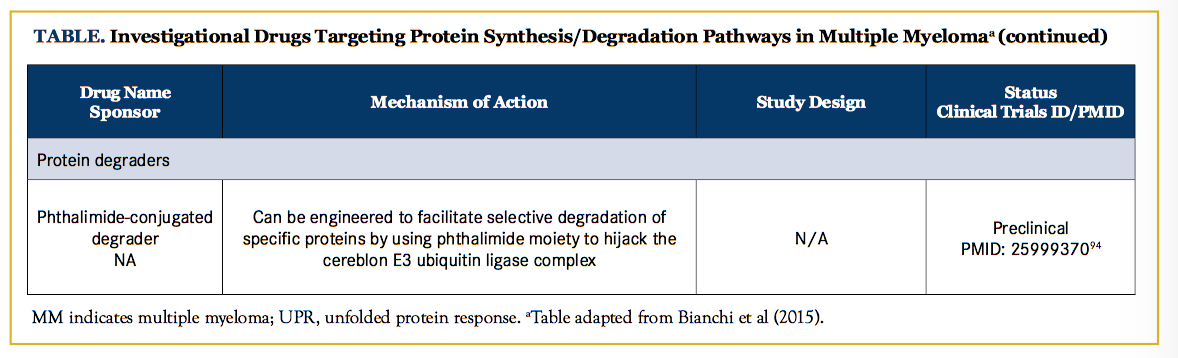
Recent insight into the mechanism of IMiDs has led to a novel therapeutic strategy (degronomid) that exploits the ability of IMiDs to redirect the cereblon E3 ubiquitin ligase complex toward specific proteins, thus targeting them for degradation.92,93 As a proof-of-concept, the phthalimide conjugate d-bromodomain and extra-terminal 1 was able to selectively induce cereblon-dependent BET protein degradation both in vitro and in mice (Figure 1F).94 This ability to hijack the UPS to selectively degrade proteins that are otherwise considered undruggable (eg, MYC, β-catenin, and myeloid cell leukemia 1) could be a powerful tool in the treatment of MM and other malignancies.
In conclusion, the understanding of MM reliance on protein- handling pathways paved the way to therapeutically target this Achilles’ heel by exacerbating baseline proteotoxic stress. In combination with IMiDs and immunotherapies, drugs targeting the protein synthesis/degradation machinery hold the key to achieving sustained remission, if not cure, in most MM patients.
Acknowledgements: We would like to thank Steven C. Smith, MD, PhD, Department of Pathology, Virginia Commonwealth University Medical Center, Richmond, Virginia, for his assistance with this report.
Financial disclosures: The authors report no relationship or financial interest with any entity that would pose a conflict of interest with the subject matter of this article
Author affiliations: Both Matthew Ho Zhi Guang and Giada Bianchi are with the department of Medical Oncology, Jerome Lipper Multiple Myeloma Center and LeBow Institute for Myeloma Therapeutics, Dana-Farber Cancer Institute, and Harvard Medical School, Boston, Massachusetts.
Address correspondence to: Giada Bianchi, MD, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215. E-mail: [email protected].


References
- Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046-1060. doi: 10.1056/NEJMra1011442.
- Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol. 2012;34(3):286-297.
- Suh DH, Kim M-KK, Kim HS, Chung HH, Song YS. Unfolded protein response to autophagy as a promising druggable target for anticancer therapy. Ann N Y Acad Sci. 2012;1271:20-32. doi: 10.1111/j.1749-6632.2012.06739.x.
- Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404(6779):770-774.
- Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett. 2007;581(19):3652-3657.
- Meriin AB, Zaarur N, Sherman MY. Association of translation factor eEF1A with defective ribosomal products generates a signal for aggresome formation. J Cell Sci. 2012;125(pt 11):2665-2674. doi: 10.1242/jcs.098954.
- van Deventer S, Neefjes J. The immunoproteasome cleans up after inflammation. Cell. 2010;142(4):517-518. doi: 10.1016/j. cell.2010.08.002.
- Yun Y, Kim K, Tschida B, et al. mTORC1 coordinates protein synthesis and immunoproteasome formation via PRAS40 to pre- vent accumulation of protein stress. Mol Cell. 2016;61(4):625-639. doi: 10.1016/j.molcel.2016.01.013.
- Selvaraju K, Mazurkiewicz M, Wang X, Gullbo J, Linder S, D’Arcy P. Inhibition of proteasome deubiquitinase activity: a strategy to overcome resistance to conventional proteasome inhibitors? Drug Resist Updat. 2015; 21-22:20-29. doi: 10.1016/j. drup.2015.06.001.
- Meister S, Schubert U, Neubert K, et al. Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer Res. 2007;67(4):1783-1792.
- Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107(12):4907-4916.
- Bianchi G, Oliva L, Cascio P, et al. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood. 2009;113(13):30403049. doi: 10.1182/blood-2008-08-172734.
- Rajkumar SV, Buadi F. Multiple myeloma: new staging systems for diagnosis, prognosis and response evaluation. Best Pract Res Clin Haematol. 2007;20(4):665-680.
- Zaarur N, Meriin AB, Bejarano E, et al. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol. 2014;34(7):1336-1348. doi: 10.1128/MCB.00103-14.
- Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272(14):9086-9092.
- Xu P, Duong DM, Seyfried NT, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133-145. doi: 10.1016/j. cell.2009.01.041.
- Kumar S, Paiva B, Anderson KC, et al. International myeloma working group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17(8):e328-e346. doi: 10.1016/S1470-2045(16)30206-6.
- Weissman AM, Shabek N, Ciechanover A. The predator be- comes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011;12(9):605-620. doi: 10.1038/nrm3173.
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550-563. doi: 10.1038/nrm2731.
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801-847.
- Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel protea- some inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103(13):1007-1017. doi: 10.1093/jnci/djr160.
- Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36(5):794-804. doi: 10.1016/j. molcel.2009.11.015.
- Mirabella AC, Pletnev AA, Downey SL, et al. Specific cell-permeable inhibitor of proteasome trypsin-like sites selectively sensitizes myeloma cells to bortezomib and carfilzomib. Chem Biol. 2011;18(5):608-618. doi: 10.1016/j.chembiol.2011.02.015.
- Demo SD, Kirk CJ, Aujay MA, et al. Antitumor activity of PR- 171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67(13):6383-6391.
- Kuhn DJ, Chen Q, Voorhees PM, et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110(9):3281-3290.
- Chauhan D, Catley L, Li G, et al. A novel orally active protea- some inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from bortezomib. Cancer Cell. 2005;8(5):407-419.
- Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15(2):122-133. doi: 10.1038/nrm3741.
- Wang X, Mazurkiewicz M, Hillert E-KK, et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci Rep. 2016;6:26979. doi: 10.1038/srep26979.
- Bianchi G, Richardson PG, Anderson KC. Promising therapies in multiple myeloma. Blood. 2015;126(3):300-310. doi: 10.1182/blood-2015-03-575365.
- Tian Z, D’Arcy P, Wang X, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706-716. doi: 10.1182/blood-2013-05-500033.
- Levine B, Kroemer G. Autophagy in the pathogenesis of dis- ease. Cell. 2008;132(1):27-42. doi: 10.1016/j.cell.2007.12.018.
- Hoang B, Benavides A, Shi Y, Frost P, Lichtenstein A. Effect of autophagy on multiple myeloma cell viability. Mol Cancer Ther. 2009;8(7):1974-1984. doi: 10.1158/1535-7163.MCT-08-1177.
- Kawaguchi T, Miyazawa K, Moriya S, et al. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol. 2011;38(3):643-654. doi: 10.3892/ijo.2010.882.
- Qiao L, Zhang J. Inhibition of lysosomal functions reduces proteasomal activity. Neurosci Lett. 2009;456(1):15-9. doi: 10.1016/j.neulet.2009.03.085.
- Lamy L, Ngo VN, Emre NC, et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23(4):435-449. doi: 10.1016/j.ccr.2013.02.017.
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143(7):1883-1898.
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524-530.
- Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3(6):388-396.
- Taylor JP, Tanaka F, Robitschek J, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet. 2003;12(7):749-757.
- Santo L, Hideshima T, Kung AL, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119(11):2579-2589. doi: 10.1182/ blood-2011-10-387365.
- De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci USA. 2006;103(16):6166-6171.
- Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551-555.
- Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36(12):2435-2444.
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631-677.
- Zhang L, Fok J, Davies FE. Heat shock proteins in multiple myeloma. Oncotarget. 2014;5(5):1132-1148.
- Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem. 2010;53(12):4585-4602. doi: 10.1021/jm100054f.
- Sabirzhanov B, Stoica BA, Hanscom M, Piao CS, Faden AI. Over-expression of HSP70 attenuates caspase-dependent and caspase-independent pathways and inhibits neuronal apoptosis. J Neurochem. 2012;123(4):542-554. doi: 10.1111/j.1471- 4159.2012.07927.x.
- Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013-1030. doi: 10.1038/nrd2755.
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081- 1086. doi: 10.1126/science.1209038.
- Woo CW, Cui D, Arellano J, et al. Adaptive suppression of the ATF4–CHOP branch of the unfolded protein response by toll-like receptor signaling. Nat Cell Biol. 2009;11(12):1473-1480. doi: 10.1038/ncb1996.
- Lee A-HH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448-7459.
- Hu MC, Gong HY, Lin GH, et al. XBP-1, a key regulator of unfolded protein response, activates transcription of IGF1 and Akt phosphorylation in zebrafish embryonic cell line. Biochem Biophys Res Commun. 2007;359(3):778-783.
- Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664-666.
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927-939.
- Davenport E, Aronson LI, Davies FE. Starving to succeed. Autophagy. 2009;5(7):1052-1054.
- Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2al- pha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14(2):230-239.
- B’Chir W, Maurin A-CC, Carraro V, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41(16):7683-7699. doi: 10.1093/nar/ gkt563.
- Carrasco DR, Sukhdeo K, Protopopova M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349-360.
-
Leung-Hagesteijn C, Erdmann N, Cheung G, et al.
Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24(3):289-304. doi: 10.1016/j.ccr.2013.08.009. - Chauhan D, Singh AV, Aujay M, et al. A novel orally active proteasome inhibitor ONX 0912 triggers in vitro and in vivo cytotoxicity in multiple myeloma. Blood. 2010;116(23):4906-4915. doi: 10.1182/blood-2010-04-276626.
- Hurchla MA, Garcia-Gomez A, Hornick MC, et al. The epoxyketone-based proteasome inhibitors carfilzomib and orally bioavailable oprozomib have anti-resorptive and bone-anabolic activity in addition to anti-myeloma effects. Leukemia. 2013;27(2):430-440. doi: 10.1038/leu.2012.183.
- Ghobrial IM, Savona MR, Vij R, et al. Final results from a multicenter, open-label, dose-escalation phase Ib/II study of single-agent oprozomib in patients with hematologic malignancies. Blood. 2016;128(22):2110-2110.
- Hari PN, Shain KH, Voorhees PM, et al. Oprozomib and dexamethasone in patients with relapsed and/or refractory multiple myeloma: initial results from the dose escalation portion of a phase Ib/II, multicenter, open-label study. Blood. 2014;124(21):3453-3453.
- Shah J, Niesvizky R, Stadtmauer E, et al. Oprozomib, pomalidomide, and dexamethasone (OPomd) in patients (Pts) with relapsed and/or refractory multiple myeloma (RRMM): initial results of a phase 1b study. Blood. 2015;126(23):378-378.
- Oerlemans R, Franke NE, Assaraf YG, et al. Molecular basis of bortezomib resistance: proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112(6):2489-2499. doi: 10.1182/blood-2007-08-104950.
- Rückrich T, Kraus M, Gogel J, et al. Characterization of the ubiquitin–proteasome system in bortezomib-adapted cells. Leukemia. 2009;23(6):1098-1105. doi: 10.1038/leu.2009.8.
- Richardson PG, Zimmerman TM, Hofmeister CC, et al. Phase 1 study of marizomib in relapsed or relapsed and refractory multiple myeloma: NPI-0052-101 Part 1. Blood. 2016;127(22):2693- 2700. doi: 10.1182/blood-2015-12-686378.
- Das DS, Ray A, Song Y, et al. Synergistic anti-myeloma activity of the proteasome inhibitor marizomib and the IMiD immuno- modulatory drug pomalidomide. Br J Haematol. 2015;171(5):798- 812. doi: 10.1111/bjh.13780.
- Tian Z, D’Arcy P, Wang X, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706-716. doi: 10.1182/ blood-2013-05-500033.
- Chauhan D, Tian Z, Nicholson B, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345-358. doi: 10.1182/blood-2013-05-500033.
- Song Y, Ray A, Li S, et al. Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia. 2016;30(9):1877- 1886. doi: 10.1038/leu.2016.97.
- Richardson PG, Badros AZ, Jagannath S, et al. Tanespimycin with bortezomib: activity in relapsed/refractory patients with multiple myeloma. Br J Haematol. 2010;150(4):428-437. doi: 10.1111/j.1365-2141.2010.08264.x.
- Seggewiss-Bernhardt R, Bargou RC, Goh YT, et al. Phase 1/1B trial of the heat shock protein 90 inhibitor NVP-AUY922 as monotherapy or in combination with bortezomib in patients with relapsed or refractory multiple myeloma. Cancer. 2015;121(13):2185-2192. doi: 10.1002/cncr.29339.
- Dimopoulos M, Siegel DS, Lonial S, et al. Vorinostat or placebo in combination with bortezomib in patients with multiple myeloma (VANTAGE 088): a multicentre, randomised, double-blind study. Lancet Oncol. 2013;14(11):1129-1140. doi: 10.1016/S1470- 2045(13)70398-X.
- Harada T, Hideshima T, Anderson KC. Histone deacetylase inhibitors in multiple myeloma: from bench to bedside. Int J Hematol. 2016;104(3):300-309. doi: 10.1007/s12185-016-2008-0.
- Yee AJ, Bensinger WI, Supko JG, et al. Ricolinostat plus lenalidomide, and dexamethasone in relapsed or refractory multiple myeloma: a multicentre phase 1b trial. Lancet Oncol. 2016;17(11):1569-1578. doi: 10.1016/S1470-2045(16)30375-8.
- Vogl DT, Raje N, Hari P, et al. Phase 1B results of ricolinostat (ACY-1215) combination therapy with bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma (MM). Blood. 2014;124(21):4764-4764.
- Minami J, Suzuki R, Mazitschek R, et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia. 2014;28(3):680-689. doi: 10.1038/leu.2013.231.
- Park HR, Tomida A, Sato S, et al. Effect on tumor cells of blocking survival response to glucose deprivation. J Natl Cancer Inst. 2004;96(17):1300-1310.
- Wang Q, Mora-Jensen H, Weniger MA, et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc Natl Acad Sci U S A. 2009;106(7):2200-2205. doi: 10.1073/pnas.0807611106.
- Uys JD, Xiong Y, Townsend DM. Nitrosative stress-induced S-glutathionylation of protein disulfide isomerase. Methods Enzymol. 2011;490:321-332. doi: 10.1016/B978-0-12-385114-7.00018-0.
- Papandreou I, Denko NC, Olson M, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311-1314. doi: 10.1182/blood-2010-08-303099.
- Ali MM, Bagratuni T, Davenport EL, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30(5):894-905. doi: 10.1038/emboj.2011.18.
- Bagratuni T, Mavrianou N, Kastritis E, Liakos C, Terpos E, Dimopoulos MA. Characterization of a PERK kinase inhibitor with anti-myeloma activity. Blood. 2015;126(23):4188-4188.
- Driessen C, Bader J, Kraus M. The HIV protease inhibitor nelfinavir: a unique oral drug that inhibits the proteasome and AKT-phosphorylation, induces ER stress and sensitizes bortezomib-refractory primary myeloma cells and primary AML cells towards bortezomib. Blood. 2010;116(21):4069-4069.
- Driessen C, Kraus M, Joerger M, et al. Treatment with the HIV protease inhibitor nelfinavir triggers the unfolded protein response and may overcome proteasome inhibitor resistance of multiple myeloma in combination with bortezomib: a phase I trial (SAKK 65/08). Haematologica. 2016;101(3):346-355. doi: 10.3324/ haematol.2015.135780.
- Bianchi G, Richardson PG, Anderson KC. Best treatment strategies in high-risk multiple myeloma: navigating a gray area. J Clin Oncol. 2014;32(20):2125-2132. doi: 10.1200/ JCO.2014.55.7900.
- Durie BG, Hoering A, Abidi MH, et al. Bortezomib with lenalidomide and dexamethasone versus lenalidomide and dexamethasone alone in patients with newly diagnosed myeloma without intent for immediate autologous stem-cell transplant (SWOG S0777): a randomised, open-label, phase 3 trial. Lancet. 2017;389(10068):519-527. doi: 10.1016/S0140-6736(16)31594-X.
- Davies F, Baz R. Lenalidomide mode of action: linking bench and clinical findings. Blood Rev. 2010;24 (Suppl) 1:S13-9. doi: 10.1016/S0268-960X(10)70004-7.
- Kronke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301-305. doi: 10.1126/sci- ence.1244851.
- Lu G, Middleton RE, Sun H, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343(6168):305-309. doi: 10.1126/ science.1244917.
- Krönke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301-305. doi: 10.1126/sci- ence.1244851.
- Winter GE, Buckley DL, Paulk J, et al. Drug development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348(6241):1376-1381. doi: 10.1126/ science.aab1433.
- Chauhan D, Tian Z, Nicholson B, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345-358. doi: 10.1016/j.ccr.2012.08.007.
- Song Y, Ray A, Li S, et al. Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia. 2016;30(9):1877- 1886. doi: 10.1038/leu.2016.97.
- Shanmugam M, McBrayer SK, Qian J, et al. Targeting glucose consumption and autophagy in myeloma with the novel nucleoside analogue 8-aminoadenosine. J Biol Chem. 2009;284(39):26816-26830. doi: 10.1074/jbc.M109.000646.
- Holstein SA, Hohl RJ. Isoprenoid biosynthetic pathway inhibition disrupts monoclonal protein secretion and induces the unfolded protein response pathway in multiple myeloma cells.Leuk Res. 2011;35(4):551-559. doi: 10.1016/j.leukres.2010.08.008.
- Cross BC, Bond PJ, Sadowski PG, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A. 2012;109(15):E869-E878. doi: 10.1073/pnas.1115623109.
- Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett. 2012;325(2):117-124. doi: 10.1016/j.canlet.2012.06.003.
- Atkins C, Liu Q, Minthorn E, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Lett. 2012;325(2):117-124. doi: 10.1016/j.canlet.2012.06.003.
- Lamottke B, Kaiser M, Mieth M, et al. The novel, orally bioavailable HSP90 inhibitor NVP-HSP990 induces cell cycle arrest and apoptosis in multiple myeloma cells and acts synergistically with melphalan by increased cleavage of caspases. Eur J Haematol. 2012;88(5):406-415. doi: 10.1111/j.1600- 0609.2012.01764.x.
- Stühmer T, Chatterjee M, Grella E, et al. Anti-myeloma activity of the novel 2-aminothienopyrimidine Hsp90 inhibitor NVP-BEP800. Br J Haematol. 2009;147(3):319-327. doi: 10.1111/j.1365-2141.2009.07852.x.
- Okawa Y, Hideshima T, Steed P, et al. SNX-2112, a selective Hsp90 inhibitor, potently inhibits tumor cell growth, angio- genesis, and osteoclastogenesis in multiple myeloma and other hematologic tumors by abrogating signaling via Akt and ERK. Blood. 2009;113(4):846-855. doi: 10.1182/blood-2008-04-151928.
- Braunstein MJ, Scott SS, Scott CM, et al. Antimyeloma effects of the heat shock protein 70 molecular chaperone inhibitor MAL3-101. J Oncol. 2011;2011:232037. doi: 10.1155/2011/232037.
- Usmani SZ, Bona RD, Chiosis G, Li Z. The anti-myeloma activity of a novel purine scaffold HSP90 inhibitor PU-H71 is via inhibition of both HSP90A and HSP90B1. J Hematol Oncol. 2010;3:40. doi: 10.1186/1756-8722-3-40.
- Okamoto J, Mikami I, Tominaga Y, et al. Inhibition of Hsp90 leads to cell cycle arrest and apoptosis in human malignant pleural mesothelioma. J Thorac Oncol. 2008;3(10):1089– 1095. doi: 10.1097/JTO.0b013e3181839693.


